Distinguishing True Autism Risk Genes from Red Herrings
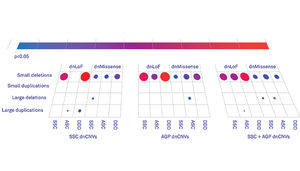
Over the past six years, gene-sequencing studies have definitively linked spontaneous (or de novo) mutations to autism in children who are the only members of their immediate family to have the condition. Studies of the Simons Simplex Collection (SSC) — a repository of data from families in which one child has autism, but the parents and siblings are unaffected — have consistently shown that the children with autism have more de novo mutations than their siblings.
In 2014, a landmark sequencing study of the protein-coding regions of the genomes of 2,515 families from the SSC identified about 400 loss-of-function mutations — ones that indisputably disrupt the function of a gene — in some of the children with autism. This doesn’t mean that all 400 mutations are in fact connected to autism, however; about 200 of the children’s siblings also have de novo loss-of-function mutations. The siblings’ mutations, though, are presumably benign, at least with respect to autism.
Assuming, as researchers believe, that the children with autism have benign mutations at about the same rate as their siblings, these numbers suggest that only about half of the 400 loss-of-function mutations in the children with autism are true autism risk genes, while the other half are red herrings. But which genes fall into which category?


