Intrinsically disordered proteins (IDPs) are a class of biomolecules with little to no stable structure. First of interest to researchers due to their role in neurodegenerative disease, these biomolecules have been found to play a role in a wide range of biological functions, challenging the sequence-structure-function paradigm. Many IDP functions are performed, not by individual proteins, but by the collective behavior of many. Indeed, phase change of IDPs has been found to underlie a host of functions like chromatin reorganization, stress granule formation, and regulation of gene expression. Using molecular dynamics simulations, we seek to unravel the molecular basis of IDP function from the individual monomer level up to characterizing the phase behavior of large condensates.
ELF3-PrD: A polyglutamine-modulated thermosensor for Arabidopsis temperature response
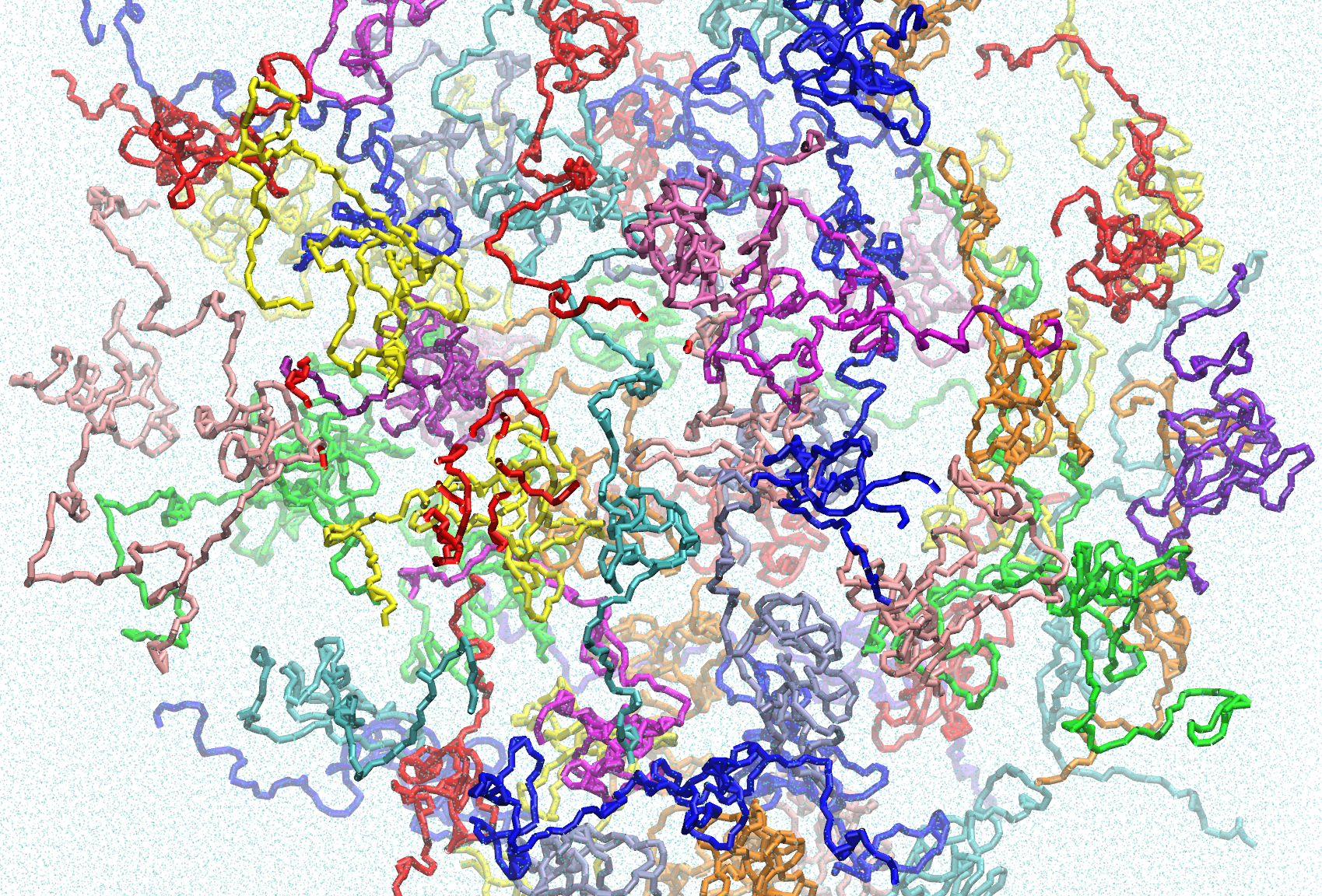
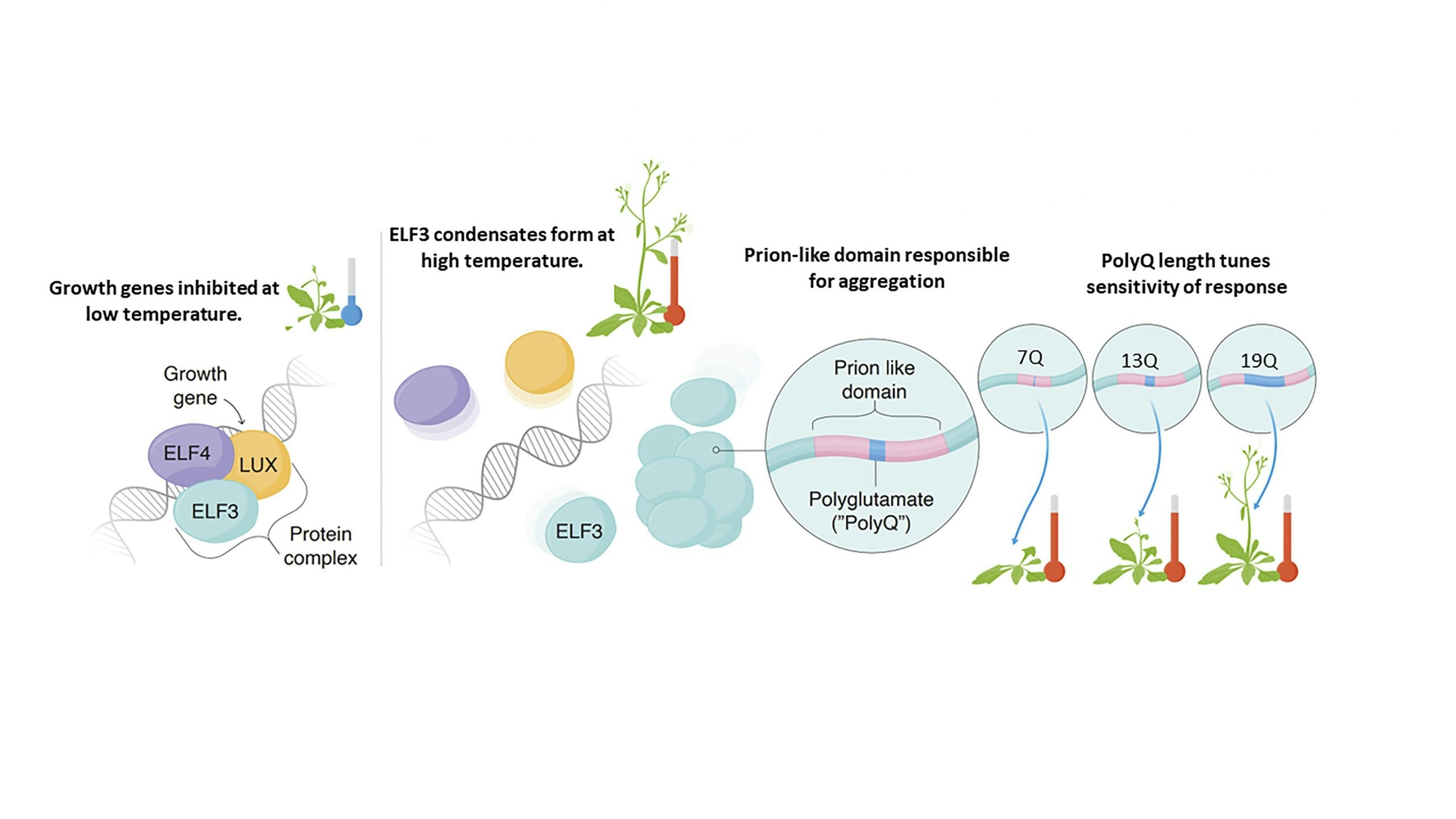
The Evening Complex (EC) allows Arabidopsis thaliana to sense and respond to environmental temperature changes. At 22°C the EC represses growth gene expression as a three protein complex. At higher temperature one EC component, ELF3, undergoes a phase change to form condensates in the nucleus, allowing transcription of growth genes to proceed. A region of low sequence complexity in ELF3, known as a ‘prion-like domain’, is responsible for aggregation, and the length of a polyglutamine tract within this region modulates the sensitivity of the temperature response. Using molecular dynamics simulations, we seek to understand the molecular mechanism of polyglutamine-modulated temperature sensing in ELF3 and the Evening Complex.
Data Release
ELF3
Hierarchical Chain growth ensembles, including 16 total ensembles comprised of 4 polyQ tract length conditions (0Q, 7Q, 13Q, 19Q) each with four temperature conditions (290K, 300K, 320K, 405K). Each ensemble contains 32,000 structures split into 64 folders, each containing 500 pdb files. The complete dataset contains 512,000 structures.
https://users.flatironinstitute.org/~ccb/elf3_data/hcg/
Data from ELF3-PrD REST2 simulations including wild-type (493,779 atoms, 505ns), F527A mutant (493,769 atoms, 506ns), and 0Q (846,088 atoms, 500ns) systems. The WT and F527A systems both have 20 replicas which are split into 8 trajectories each. The 0Q system has 12 replicas, each with a single trajectory. For each system, processed trajectories of 500ns without water and ions are provided for replicas representing 290K, 300K, 320K and 405K for easier analysis.
Publications
Richard J. Lindsay, Philip A. Wigge, Sonya M. Hanson. 2023. Molecular basis of polyglutamine-modulated ELF3 phase change in Arabidopsis temperature response. bioRxiv doi: https://doi.org/10.1101/2023.03.15.532793