A Software, a Community and a Different Way to Do Science
To answer the most complex questions in science — the nature of life, the origins of the universe — scientists often develop their own software programs to help solve their labs’ most novel and pressing problems. This type of software is ad hoc in nature, often growing organically in bits and pieces, written in different computing languages to solve an instant need, by researchers with little thought of broadening the software into something that all labs can use.
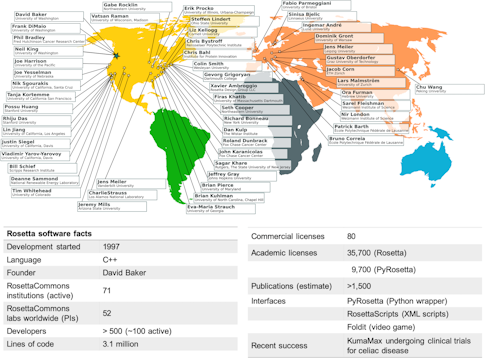
Rising up against this computational tower of Babel is Rosetta, a suite of software tools for macromolecular modeling and design. Like its namesake, the Rosetta stone, which gave the modern world a key to deciphering ancient hieroglyphs, Rosetta was first intended as a key for deciphering proteins, the building blocks of life. Designed originally to predict individual protein structure, the software has broadened in scope: It can now help scientists map complex interactions between proteins and design novel proteins. It can also boost a whole host of other biological applications in fields from medicine to synthetic materials to climate science. With 500 developers at over 70 academic institutions worldwide, Rosetta is defined not only by what it does, but also by a community of scientists who are changing how science is done and how collaborations thrive and move science forward.
—
“Rosetta was born in the wild, the raw and the unstructured,” recalls Richard Bonneau, a group leader for systems biology at the Flatiron Institute’s Center for Computational Biology. As a student in David Baker’s biochemistry lab at the University of Washington in the mid-’90s, he and several members of the lab sat down to write a code that would predict protein structure — solving a problem that had long eluded researchers. With 3.1 million lines of code and over 35,000 licenses, the Rosetta of 2020 looks very different from the one Bonneau helped craft 25 years ago. What remains the same, however, is the intent to build a standardized, shareable code that anyone can use, and to grow a cohesive community to further evolve and strengthen the code base.
“David Baker had a view early on that this community would meet regularly and that the code would be centralized,” says Roland Dunbrack, a Rosetta principal investigator and a professor in the Molecular Therapeutics Program at the Fox Chase Cancer Center.
—
Our knowledge of biology has transformed over the last few decades, but the fundamental relationship between a molecule’s structure and function is still a guiding principle of discovery-driven biological research. Rosetta assesses the structure of proteins and other biological molecules — whether natural or designed — by considering all aspects of a molecule’s conformation, from how the individual atoms attract or repel each other to how segments of a molecule can move freely in space. It then selects the structure with the lowest free energy. This information is critical for scientists working to decipher protein structure and function. Recently, improved structure prediction and a burst of new applications have ballooned Rosetta’s offerings to include over 80 distinct methods for macromolecular modeling, as reported on June 1 in Nature Methods — a milestone that represents a boon to the scientific world.
Communicating all of Rosetta’s capabilities is one of the many challenges of managing a colossal software suite and a community of thousands of users. The recent Nature Methods paper is an important step toward that goal, however, serving as a catalog for the community of Rosetta users and the larger scientific community, says Julia Koehler Leman, one of the paper’s first authors and a research scientist in systems biology in Bonneau’s group at the Flatiron Institute. With over 100 authors, the paper reviews Rosetta’s advances over the last five years, with an emphasis on major scientific applications, user interfaces and usability.
The Nature Methods resource also highlights Rosetta’s approach to several unique challenges to modeling and understanding in biology. Take membrane proteins, which are targets for 60% of the pharmaceuticals on the market despite making up just 30% of all human proteins. Because they are hard to work with in the lab, they make up a tiny fraction of the proteins available in structure databases, which Rosetta uses for its prediction algorithms. An additional obstacle is that Rosetta was developed for proteins in water rather than those embedded within cell membranes, which are ‘greasy’ and water insoluble. As a postdoctoral fellow who had also worked in an experimental membrane proteins lab during graduate school, Koehler Leman worked with colleagues to adapt Rosetta to the membrane environment. “The training I had experimentally with membrane proteins shaped how I develop code,” Koehler Leman says, and led her to emphasize ease of the user interface in her coding. Rosetta now offers an array of capabilities for modeling the characteristics of membrane proteins, including protein-protein docking and design.
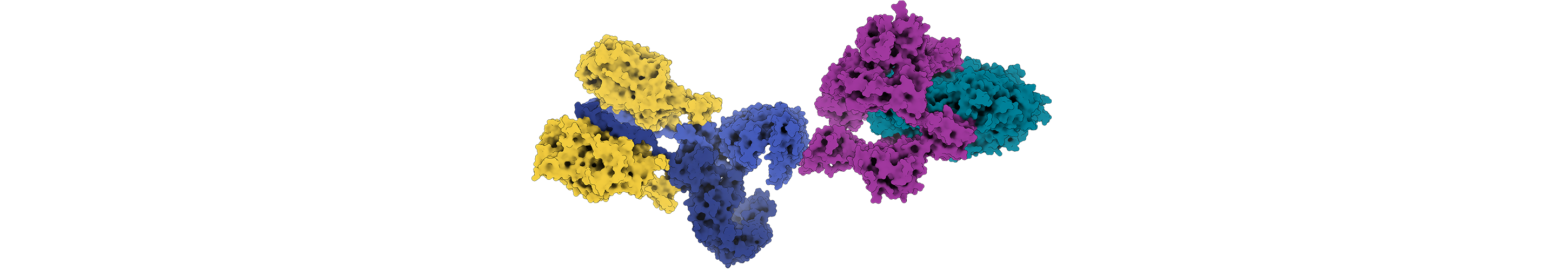
Antibodies, the proteins of the immune system, are another challenge for Rosetta. Unlike other proteins, they contain loop regions that can confound structure prediction. They are also known to make split-second changes when binding to an antigen, making them difficult to predict and model. A large collaboration of researchers, including Jeffrey Gray, a Rosetta principal investigator and a professor of chemical and biomolecular engineering at Johns Hopkins University, has succeeded in creating Rosetta methods to predict the structure of an antibody from its sequence, and then model the interaction of the antibody with its antigen. Understanding these interactions is critical for developing therapeutic antibodies or vaccines. Motivated by COVID-19, Gray, Dunbrack and other Rosetta developers are thinking about how to most effectively design antibodies to combat this and future pandemics. “Our collaborations through Rosetta have given us deep internal knowledge of antibodies,” says Gray. “The synergistic and positive nature of this community has helped us accelerate science.”

Rosetta has expanded beyond proteins, to RNA and DNA. RNA structure in particular presents challenges distinct from those of proteins. Loops with irregular nucleotide pairing abound, and the method Rosetta uses for proteins flounders in the presence of RNA; multiple possible energy minima can confound the overall energetic view of a conformation, much the way deep potholes on a hill might mislead an altimeter. Rosetta developers have demonstrated RNA structure prediction, as well as RNA- and DNA-protein binding, by modeling the molecules in a step-by-step fashion, in essence sacrificing computational expense for accuracy. Several of the leading COVID-19 vaccine candidates, including two of those selected for Operation Warp Speed, initiated by the federal government to accelerate vaccine development against COVID-19, are DNA- or RNA-based. This underscores the importance of making tools available to probe nucleic acids and how they bind to proteins.
Rosetta’s modular nature is its secret weapon: Scientists can build a dizzying array of workflows from the thousands of available code classes. “There are things we can do with Rosetta that we can’t do otherwise, like design proteins so stable they are more like nonliving materials and integrate high-throughput computation with high-throughput experiments,” says Bonneau. The sheer size of both the software itself and the worldwide community can, at times, feel unwieldy, added Bonneau, but ultimately it is necessary for solving big scientific problems.
Rosetta’s licensing agreement is unique in that most of the fees paid by pharmaceutical companies flow back to the RosettaCommons, the community of developers, to support code maintenance and community building. “You can think of Rosetta as a multi-institution research group, with money,” says Dunbrack. “There are lots of consortiums out there, but not as many with their own source of funds.” Recently, the corporate licensing agreement was changed so companies can contribute code back to Rosetta. “This change says a lot about where our tools are, and how the community and the science are evolving,” says Brian Weitzner, another of the Nature Methods paper’s first authors and a senior scientist at Lyell Immunopharma, a company Baker co-founded.
Maintaining the code takes great effort and coordination. Each time a developer submits a piece of code, it has to be integrated into the entire Rosetta suite. “Individual code development branches are merged into the software several times a day,” says Koehler Leman, “so we need to continually test the software to make sure it won’t break.” The benefits make this effort worthwhile, says Bonneau. “For whatever you want to do, whether with DNA, RNA, drugs or surfaces, you might just have two to 10 people in the community writing a code in the same framework.” RosettaCommons issues its own grants to members of the community for code maintenance, something scientific research grants won’t often cover.
The emphasis on documentation and interface development aims to make Rosetta more user-friendly and a benchmark for how people can develop powerful software in any community, says Koehler Leman. Detailed user instructions, called protocol captures, accompany each new addition of code, and three different language interfaces (C++, Python and command line) are available to developers. For the general public, including K-12 students, the video game Foldit offers a chance to play with protein structure, with terms like ‘rubber bands’ for restraints and ‘shake’ for rotating parts of a molecule. Foldit’s 700,000 regular users routinely solve real-world scientific structure puzzles, including a challenge this past February to design a protein to inhibit the spike protein on the new coronavirus, with the top results selected for experimental testing in labs.
To rally the community around the arduous tasks of standardized documentation and code curation, RosettaCommons holds a meeting (RosettaCon) each summer and winter, hack-a-thons for code maintenance and improvement, and boot camps to train junior developers. It also grants an annual Rosetta Service Award for contributions to code maintenance or community leadership. A conversation in 2012 between Weitzner and Andrew Leaver-Fay, now an assistant professor of biochemistry and biophysics at the University of North Carolina School of Medicine and Matthew O’Meara, now a research assistant professor of computational medicine and bioinformatics at the University of Michigan Medical School, led to the creation of the boot camp. “We noticed postdocs spent a year and a half learning to program, and we said, let’s have a class. I’ve learned that when you ask, ‘What if we did this differently?’ the community is so supportive and the response is, ‘Yeah, let’s do it,’” says Weitzner, who worked in Dunbrack’s lab in high school and college, in Gray’s lab in graduate school, and in Baker’s lab as a postdoc.
In an internship program, college students can spend a summer in a Rosetta lab, sandwiched between a week at the Coding Boot Camp at the University of North Carolina and a week at the summer RosettaCon in Washington state. “They start to foster this community right when you come in,” says former intern and current Coding Boot Camp teaching assistant Anna Yaschenko, who graduated from the University of Maryland this year with a dual major in computer science and bioinformatics. “RosettaCon is so casual — it allows people to connect in ways you couldn’t at a typical conference. I was surprised at how tight-knit the community was despite being so large.”
A post-baccalaureate program starts this summer, Gray says, and all five participants are from groups underrepresented in STEM fields. Rosetta’s diversity, equity and inclusion committee has encouraged Rosetta principal investigators, students and postdocs to attend conferences like the Annual Biomedical Research Conference for Minority Students; oSTEM, a professional society for LGBTQ people in STEM fields; and the Grace Hopper Celebration of Women in Computing conference. “Diversity in research programs is important because it’s fair, and everyone should have the opportunity to participate,” says Dunbrack. RosettaCommons recently put out a statement on Black Lives Matter that included action items for individuals and labs to combat racism. “We had so many people weighing in,” says Gray, “saying ‘this is important,’ or ‘here’s this subtlety.’ There’s still a lot of work to do, but I was very proud of our community for their serious engagement.”
As a software and a community, Rosetta represents a different way to do science. “We really believe the best idea wins, no matter where it comes from,” says Koehler Leman. “At our conferences, people are less worried about being right or wrong, and more concerned with ‘Does something work or not?’”
“Other research communities can benefit from this approach,” says Weitzner, “and collaborate more and not worry as much about competing.” The challenge will be to continue to balance innovation with standardization as the software and community grow. “We’ve got to maintain the quality and continuity of the code, while integrating new methods and research into Rosetta,” says Bonneau. “New problems in biology have a scale and complexity that demand this kind of collaboration.”


