Our Cells Often Behave in Unpredictable and Unknown Ways
Molecular biologist and Simons Junior Fellow Naama Aviram works to understand how individual cells function — a more mysterious process than often appreciated. Beyond the sheer lure of scientific discovery, Aviram argues better understanding how cells work could lead to new treatments for complex illnesses.

Simons Junior Fellow Naama Aviram thought she would spend her life studying marine ecosystems. In the end, she was drawn to the drama of how cells build and sustain life. In biology textbooks, the explanation of how cells work is too simplified and tidy because many details about these cellular processes remain unknown. Aviram, a molecular biologist, is working to shed light on these processes. Previously, her research helped to map the organization and position of proteins within cells, and she is currently interested in learning how some of the simplest organisms in existence — bacteria — respond when they encounter a virus. By using this knowledge to harness the natural immune properties of bacteria, this line of research could lay the foundation for new treatments or cures for debilitating illnesses.
Aviram holds both a master’s and a doctoral degree in molecular genetics from the Weizmann Institute of Science in Israel. Now a postdoc in Luciano Marraffini’s lab at Rockefeller University, she has won several awards for excellence in science, including the Israel National Postdoctoral Award for Advancing Women in Science.
Aviram and I recently discussed the lure of molecular biology, what she has learned so far and what she hopes to do next. Our conversation has been edited for clarity.
What drew you to molecular biology?
My undergraduate major was marine biology, and I thought I would spend my life studying nature preservation and the ecology of the sea. I love the sea, and I’ve been scuba diving since I was 12 years old.
When I began college, I didn’t know much about the smaller scale of biology — the cellular and molecular biological processes that underlie all of life. But once I was introduced to it, I was hooked. The fact that a single atom travels from one molecule to another, and that the accumulation of these journeys is at the heart of what sustains life on earth, was amazing to me. I knew this is what I was supposed to study.
I see my work as solving cellular mysteries — with one crucial caveat. In science, the discoveries we make bring us closer to a fuller and more complete understanding of biology or whatever our field of interest might be. In my case, that is the molecular processes happening inside cells. But no scientific discovery is ever final; instead, we accept explanations as long as the data supports it, and then evolve that understanding as new data emerge. And in my area of focus, there are always new data — and new insights — emerging.
I thought we already understood many of the crucial processes in cellular biology. But it sounds like that’s not the case.
Yes, there’s a lot we still don’t understand, even though we might think we do.
If you open a biology textbook, you’ll find a depiction of a cell. That picture might show different parts of the cell, like mitochondria (which help the cell produce energy) and the endoplasmic reticulum (which is sort of like the cell’s post office, shuttling proteins to the right place). This picture makes it seem like everything is tidy and organized.
The reality is that cellular processes are messy and dynamic, and we can’t always say how these processes work. For example: How do proteins move from one part of a cell to another, or between cells? There’s a lot we don’t yet know. During my doctoral work at the Weizmann Institute of Science, I investigated a specific part of this conundrum: how some proteins get from point A to point B to execute their function.
Interesting question. Tell me more!
When I began graduate school, scientists in the field knew about two main protein-targeting pathways. One, discovered in the 1980s, is known as the SRP pathway. My doctoral advisor Dr. Maya Schuldiner discovered another more recently, called the GET pathway. Each of these pathways caters for a different subset of proteins destined to end up in the endoplasmic reticulum, that cellular ‘post office’ I mentioned earlier.
However, scientists in our lab and elsewhere also knew that if you disrupt these pathways — deliberately making it hard for the proteins to reach the endoplasmic reticulum — about half of those proteins arrived anyway. This told us that another, undiscovered, pathway or process was also in use.
In 2016 we published a paper in Nature showing in yeast that certain factors — which we called SND (SRP-independent) factors — leap into action in situations where a protein is not recognized by the SRP or GET pathways. Essentially, SND factors served as an alternative targeting pathway; they can also serve as backup when the other two pathways aren’t working. Though there may be more pathways like this yet to be discovered, this work felt like a significant advance in cell biology.
What else did you discover during your doctoral research?
In addition to these protein-targeting pathways, I also studied what happens after a molecular ‘mistake,’ when proteins arrive to the wrong part of the cell. Let’s say a protein is meant to go to the mitochondria but for some reason ends up in the endoplasmic reticulum. It turns out that cells appreciate the danger of this situation. They’ve developed machinery that extracts proteins from the wrong spot and gives them a second chance to get to the right place. Working with colleagues at the Weizmann Institute and the University of Kaiserslautern in Germany, we identified this process by using a combination of genetic manipulations, fluorescent microscopy and classical biochemistry assays. We published this work in Science in 2018.
This correction mechanism is important because if the protein stays in the wrong place it could lead to cell toxicity or cell death. Just as with discovering a new protein pathway, finding this protein retrieval mechanism felt like a significant advance. There may be therapeutic applications that could build upon the cell’s natural corrective processes.
That all sounds very gratifying! How has your postdoctoral work as a Simons Junior Fellow at Rockefeller changed from your graduate school days?
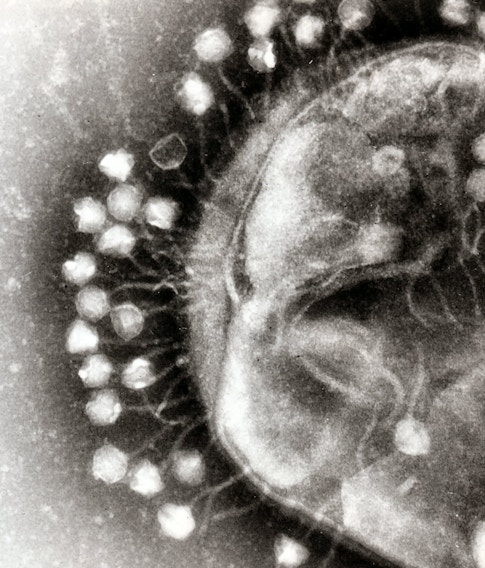
These days I am focused on CRISPR, which many people probably know as a gene-editing tool that enables very precise changes to genes. Though many complex ethical questions surround its applications, CRISPR genome-editing is a significant technological advance, and it’s no surprise that its developers earned the Nobel Prize in Chemistry last year. As someone who was in biology before and after the so-called ‘CRISPR revolution,’ I saw the excitement around this spectacular advance firsthand.
However, it is important to note that before it was developed as a gene-editing tool, CRISPR was originally a group of adaptive immune systems that naturally exist in prokaryotes, simple organisms like bacteria. We know that CRISPR-harboring bacteria can acquire immunological memories of viruses that they encounter. Then, in case of reinfection by the same virus, bacteria use these memories to snip out the genome of the virus before it can cause any new damage. The thought that a single bacterial cell can acquire long-term immunological memories is fascinating to me! The process of acquisition of immunological memories, meaning the regions of the virus that are being ‘learned’ by the bacteria, are not random. However, we do not fully understand how or why bacteria choose these exact regions. My hypothesis is that certain regulatory elements within the cell, which may correlate to what the cell senses during viral infections, serve as cues for forming immunological memories. This hypothesis is what the Simons Fellowship is allowing me to study, and it is ongoing work.
What do you believe you have most gained from your Simons Fellowship?
The main benefit of the fellowship, besides the very generous financial support, is the chance to interact with experts in virtually any field of science you can think of.
Prior to the pandemic, I went to as many fellow dinners as I possibly could, because the presentations and conversations with other Junior Fellows were so stimulating. I will certainly take the intellectual stimulation and community-building inspired by the fellowship with me throughout my career. I am also thankful to the Simons Foundation for extending my fellowship for an extra year, due to pandemic-related delays in my work, which is enabling me to complete my research and help me lay the foundation for the next phase of my career.