Systems Neuroscience Is About to Get Bonkers
We are all familiar with the standard paradigm in systems neuroscience: Study a simplified behavior in a model organism while measuring neural activity in brain structures thought to subserve the behavior. Analyze responses from a single neuron, or more recently, a population of neurons in one area, and correlate those responses back to behavior.
But have you ever wondered what systems neuroscience will look like when every single laboratory is recording from many thousands of neurons simultaneously? The potential of these technologies is exciting — we finally have the opportunity to determine if we can understand how populations of neurons give rise to the computations that lead to behavior. But if we are to have a hope of answering that question, we need to start thinking through how we will manage the flood of data such technologies will produce. Without theoretical frameworks and computational tools, neuroscience will drown in the deluge.
Two advances are making the impending data surge an important issue to address now. The first is the advent of widely available, low-cost recording technologies that are easy to use, such as the Neuropixels probes. These probes promise high neuron count, high temporal resolution measurements that experimentalists can quickly and easily deploy in a wide number of animal models. They can be combined with optical imaging to study neural activity across the brain and over time.
The second is the maturation of deep learning, a catchphrase for a collection of very powerful artificial neural network concepts, along with the software and hardware that power them. Driven by algorithmic innovation and computing advances over the last decade, deep learning has emerged as an important framework for modeling huge quantities of high-dimensional, time-series data.

Making progress without a theory
Technologies like Neuropixels make it possible to address the big question in systems neuroscience: how do large populations of neurons give rise to behavior? But we currently have no useful theory of biological neural computation to organize our thinking about this question or to guide the design of new experiments. Yet, in the absence of such a theory, we still need to think about the most important measurements to make.
Here are some possible first steps:
- Do what we do now, but record from more neurons across more brain regions. Researchers are already using technologies such as Neuropixels to measure brain-wide responses to simple, well-studied behaviors. These types of studies will give insight into how neurons in different brain regions work together to perform computations.
- Do what we do now, but make the tasks we study more complex. To capture the brain’s full computational repertoire, we may need to use much more complex behavioral tasks. Of course, more complex tasks will also produce more complex data, which will in turn be more challenging to interpret.
- Systematize. We could also focus on previously understudied brain structures. So much of the brain is essentially uncharted territory. Recording methodologies that increase our understanding of population responses in critical structures such as the thalamus, basal ganglia or cerebellum will be of immense importance.
Deep learning as a powerful tool for neuroscience
These types of experiments will produce huge volumes of data, but the field generally lacks sophisticated ways to analyze simultaneous recordings from thousands of neurons. As a computational neuroscientist, I have focused on developing deep-learning tools to help neuroscientists make sense of this data.
In analogy to biological neural networks, artificial neural networks (ANNs) are composed of simple nonlinear elements, they are potentially recurrently connected, and the synapses that connect the units determine the computation. The gross structural similarities between artificial and biological neural networks provide some justification that ANNs may provide insight into the functioning of biological networks. However, further theoretical work is needed here to understand what architectural details of an ANN better align its population activity with that of a biological neural network.
One successful deep-learning approach involves modeling behavioral tasks. Researchers build an ANN and optimize it to solve a task analogous to the one studied in animals. They then compare the internals of the trained ANN with the biological neural recordings, typically smoothed spike trains of a population of neurons. If there are quantitative similarities, researchers can then attempt to reverse-engineer the ANN in order to develop a mechanistic explanation for how the ANN solves the task. The insights found in the ANN can lead to testable hypotheses for how the biological network implements the behavior. For example, we first used this approach in 2013 to understand how the brain might implement a context-dependent sensory integration task. Others are using neural networks to study visual processing.
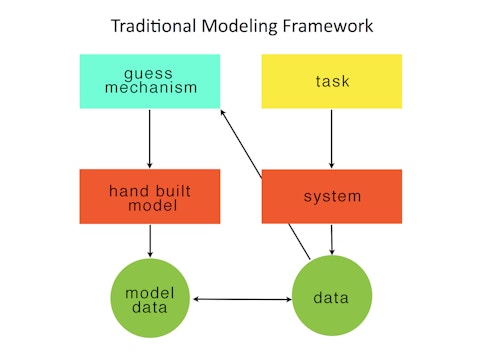
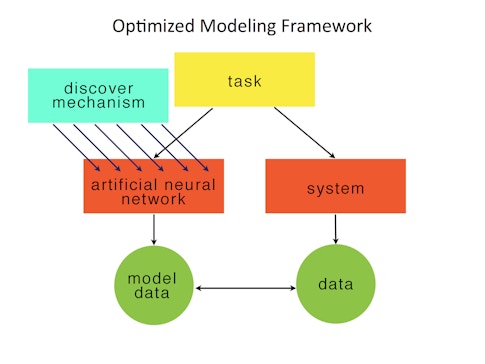
Modeling the task is an important use of deep learning in neuroscience because it is a form of automated hypothesis generation. One assumes that a strong task specification will lead to solutions that can provide the backbone for a theory of how a particular neural computational subserves a behavior. An implicit assumption is that optimized models can be reverse-engineered or in some other way be more easily understood than the biological circuits. This assumption may not be true. But at the very least, one has the entire circuit and can build many, many slight variations for systematized study. I think the systems neuroscience community should think of ANNs trained to model tasks as a kind of synthetic model organism.
Several other deep-learning and machine-learning advances are also yielding fruit:
- Model the data: Optimize a neural network that generates simultaneously recorded neural data. Modeling the data is useful because it provides a concrete network that may give insight into how the neural data is generated. Additionally, modeling the data is often used to find meaningful, concise representations of the data. For example, we developed latent factor analysis via dynamical systems (LFADS) to discover latent temporal factors in single-trial neural data. Another technique uses switching linear dynamical systems to build in a more interpretable dynamics to generate neural data.
- Modeling behavior: New automated methods for tracking and analyzing behavior (in some cases simultaneously with neural activity) generate new insight into the structure of behavior. Precise quantitative measurement of behavior will be critical to mitigate the complexity of high-dimensional neural data.
- Connectomics: New algorithms are dramatically speeding the ability to generate wiring diagrams of the brain. More detailed connectomics diagrams may shed light on circuit motifs that generate the dynamic neural patterns that subserve behavior. The ring attractor network in the fly provides an example of how circuit structure can inform dynamics and vice versa.
Along with optimizing models, we will surely continue to build models by hand as well as developing word-based explanations of our data. More broadly, are there any theoretical or conceptual frameworks outside of machine learning that can treat this kind of high-dimensional data? Perhaps we will find leadership in statistics. If so, which statistics should we concern ourselves with, and what would they tell us? One example may be statistics of how task complexity affects the number of measurements we need to make in high-dimensional data. Finally, physicists have traditionally applied statistical mechanics to high-dimensional data. However, statistical mechanics typically makes strong assumptions about randomness in the data. Will these assumptions hold in large neural recordings?
Many researchers are already working on essentially all of the topics I mentioned above. My guess is that even if it takes a long time to formulate useful experimental programs and modeling studies to answer our “grand” question, these kinds of measurements and models will still enable us to make progress by offering a set of emergent principles that tend to crop up under common circumstances. Some examples include linear algebra concepts such as null spaces or the use of common dynamical systems concepts such as the systematic organization of initial conditions and inputs in a lawful state-space. Even if these motifs do not rise to the level of a theory, they may nevertheless help us organize our thinking about how neural populations give rise to behavior.
The need for collaboration
It appears to me that the do-it-yourself, cottage-industry perspective currently pervasive in neuroscience will have to change. The systems neuroscience I am envisioning will eventually require large collaborations of experimentalists, engineers and theoreticians to handle the theoretical, computational and practical realities of large-scale data collection and analysis. It will ultimately require rethinking how neuroscientists organize themselves and how resources and credit are distributed across efforts that are truly multidisciplinary. For example, the International Brain Laboratory, a collaboration that spans 21 labs and is funded by the SCGB and Wellcome Trust, serves as a test bed for large-scale collaboration in neuroscience.
As a serious practical matter, researchers will need to save, curate and manipulate enormous quantities of data going forward. These datasets will be too large for any one group to fully understand. Data sharing and open-source formatting standards, such as Neurodata Without Borders and DataJoint, will be of paramount importance, enabling researchers to collaborate freely and allowing specialist expertise meaningful access to data. If the field of deep learning is any indication, open-source, easily available datasets will be the drivers that move the field forward (e.g., MNIST, TIMIT, or ImageNet).
What can students and currently practicing scientists do to prepare? The difficulty here is that we don’t know exactly which skills and knowledge will turn out to be important. My guess is that many future systems neuroscientists will be hybrid experimentalist/modeler/engineers and some will be specialists. Both theorists and experimentalists can benefit from immersing themselves in the other’s culture.
- Theorists, modelers and engineers should consider a postdoc or sabbatical embedded in an experimental lab. This does not necessarily mean doing experiments, but rather to become fluent in the data, techniques and language of a given lab. As a computational neuroscientist, embedding in the lab of Krishna Shenoy, an experimentalist, was an enormously formative experience, as important as getting my Ph.D.
- Experimentalists should enroll in computational summer schools tailored to neuroscience. They should consider taking classes in the following topics, which are proving to be useful from a theoretical perspective: deep learning, linear algebra, probability and statistics, linear systems theory (as taught in electrical engineering), nonlinear dynamical systems, calculus, and programming classes. If appropriate, they should also consider a postdoc or sabbatical in a theoretical/modeling group.
In summary, systems neuroscience is about to get bonkers. The advent of inexpensive, high-quality recording equipment with the maturation of deep learning is an incredible opportunity! There couldn’t be a better time to be in the field, but we should be preparing now.
David Sussillo is a research scientist at Google in the Google Brain group. Sussillo is also an adjunct faculty member in the Electrical Engineering Department at Stanford University and an investigator with the Simons Collaboration on the Global Brain. The views above do not necessarily reflect those of Google.


