
The Vast Viral World: What We Know (and Don’t Know)

Slightly ovoid in shape and somewhat blurred at the edges, the black splotches were scattered across a mottled gray background, looking much like a postmodern painting. At a meeting of the Medical Society of Berlin in 1938, Helmut Ruska, MD, a German physician and biologist, was presenting the first images ever seen of virus particles. The splotches were members of the poxvirus family, specifically ectromelia, visualized directly in the lymph fluids of infected mice.
Thanks to the invention of the electron microscope, then known as the Übermikroscop, scientists could finally observe—actually see—what was already known to exist: the minuscule and mysterious world of viruses. Ruska’s brother, Ernst, a physicist, had built the first prototype of the instrument while completing his PhD, but Helmut saw the device’s potential application to the field of biology. Helmut and his colleagues went on to gather nearly 2,000 black-and-white images by the end of 1939. Their collection included a variety of pathogens known to infect humans and plants, including the variola virus (which causes smallpox) and tobacco mosaic virus (the first virus ever discovered).
Just over 80 years later, scientists published the first images of SARS-CoV-2, the novel virus that causes COVID-19. To reveal its iconic spike protein, they used an instrument similar to Ruska’s. The black-and-white images visually confirmed that this was a member of the coronavirus family, enabling scientists around the world to begin drawing upon existing knowledge of such viruses from years of prior research. Scientists were able to directly observe this new virus soon after a cluster of patients in China was diagnosed in late December 2019 with a pneumonia of unknown cause. By March 2020, COVID-19 had been declared a pandemic by the World Health Organization.
Events moved far more slowly in previous pandemics. The Black Death pandemic in the mid-1300s killed more than one-third of Europe’s population in four years, without anyone knowing what caused it. By the time of the 1918 flu pandemic, scientists understood the disease’s cause but couldn’t see the virus or test for it. When SARS-CoV-2 emerged, the value of basic science research conducted many decades earlier became quickly evident in the form of innovative tools for virus detection. Researchers can now sequence novel viruses and study their genomes, but even so the electron microscope continues to offer a unique “open” view of such pathogens.
“You can’t really argue with a picture,” says Cynthia Goldsmith, who uses electron microscopy to identify pathogens for the U.S. Centers for Disease Control. “This technology allows us to see not only how the virus looks but also how it grows in the cell.” Although Goldsmith has identified so many viruses over the years that she can’t even guess the number, the viruses she’s examined are limited to those already known to cause disease—and those that primarily infect people. But scientists estimate that 1.67 million yet-to-be discovered viral species exist in mammal and bird hosts. Bats, for example, teem with new strains of coronaviruses. There may be more viruses than there are stars in the universe. Basic science research being conducted today will play a critical role in helping us to understand their past, present, and future—and better prepare us for the emergence of still more novel viruses.
“Scientists and public health officials have spent a lot of time thinking about when and where the next pandemic will emerge,” says Laura Bloomfield, a recently minted MD-PhD from Stanford University. “But what we’ve learned is that the likelihood of predicting that precisely is very low. We need to put the structures in place to prepare and respond but also to prevent future outbreaks in the first place.”
That will mean engaging in more basic science research. It will mean not only discovering and sequencing more viruses, but also investing in research that will improve our ability to understand the structures of viruses, how they interact with their hosts, and how they have evolved through history. In addition, preparing for the next pandemic will also mean focusing more on human behavior and repairing our relationship with nature. Activities such as clearing native forests for industrial forestry or agriculture, expanding human settlement or land use into intact ecosystems, and trading wildlife greatly increase the chances of another catastrophic outbreak.
The discovery of viruses
In Ruska’s day, researchers used the term “filterable infectious agents” to describe viruses. Experimentation had revealed that these tiny entities, invisible to the naked eye, were capable of passing through filters that caught and retained bacteria and other microorganisms. Coined from a Latin word meaning “slimy liquid,” “potent juice,” or “poison,” the term “virus” was applied to agents both tiny enough to pass through filters and unable to reproduce without the aid of a living host—plant or animal. Martinus Beijerinck, DSc, a Dutch microbiologist and botanist, demonstrated that viruses cause disease by experimenting with sap from sickly tobacco-plant leaves during the 19th century.
He was one of the first scientists to show that an entity too small to be detected by a light microscope is capable of infecting plants. Beijerinck’s application of freshly extracted sap from sick plants produced spots on the leaves of healthy plants; the more sap he applied, the worse the symptoms—including malformed leaves. He called the sap from the sick plants contagium vivum fluidum (contagious living fluid). He noted it was “drawn into the growth of dividing cells,” meaning it required other life (plant cells, in this case) to reproduce.
Beijerinck’s discovery of this liquid poison capable of infecting other plants laid the foundation for our ability, today, to identify new viruses, decipher their morphology (their structure), and use this information to develop treatments against the diseases they cause. But it also biased our popular perception of viruses as solely pathogenic—that is, capable of causing illness. In most cases, viruses don’t cause disease, and many are beneficial. They shape our life, for better or worse, with some viruses posing health threats but many others serving helpful functions.
How viruses shape life
Simple in structure but advanced in function, viruses exist in a category that lies between the inert and the living. Some biologists think of them as quasi-alive, while others refer to them as inanimate tiny particles or arrangements of matter. But experts agree they are like vines wrapping around the tree of life; their existence involves symbiosis, an intimate association between two forms of life. Viruses can reproduce only inside living cells. Even at our healthiest, our bodies are home to trillions of viruses, maybe more. There, they are most “alive,” as their genetic material penetrates into their host’s cells and turns them into virus-making factories.
“Think of a virus and a cell together as a microscopic copy machine,” explains Sarah Olson, PhD, a disease ecologist with the Wildlife Conservation Society’s Health Program. “The virus uses existing entry points to get into the cell—and when it enters, it’s carrying instructions to take over the cell’s machinery and make copies of itself. Then off it goes to the next.”
A virus replicates through a series of steps, first attaching to a host cell and injecting its genetic material, then incorporating itself into the cell’s genetic material as the cell churns out more viruses. Once the viral information becomes integrated into a host’s genome, that information becomes part of the host and can be passed across generations. Indeed, approximately 8% of the human genome comes from viral genes, which means we’ve evolved along with viruses and will continue doing so. In fact, without viruses, humans and other mammals might still be laying eggs. At some point during evolution, the protein syncytin, which is essential for the formation of the placenta, came from a retrovirus infection. We also know that viruses can keep bacteria from invading our gut and causing infections.
Like other living creatures, viruses also carry out a Darwinian mission: They aim to increase in number as much as possible, to extend their geographic boundaries, and to survive across time. That is exactly what SARS-CoV-2 is doing now, even as vaccines roll out and new variants are identified. But the past is still important. As Michael Emerman, PhD, a biologist at the Fred Hutchinson Cancer Research Center in Seattle says, the motivation for studying ancient events in the evolutionary history of viruses is to better understand pressing modern challenges.
The origins of viruses and why their history matters
Patterns that have emerged from basic research on the origins of viruses and their evolution may help inform our preparedness for future pandemics. “You’d like to have a single explanation for the origins of all current viruses,” says Julia Durzyńska, PhD, a biologist at Adam Mickiewicz University in Poznań, Poland, who has written about viral evolution. “Instead, there are three dominant models that seem to complete each other.”
The so-called “virus-first” model suggests that at the dawn of life, very simple forms of viruses existed before cells. The ancient ancestors of modern viruses may have provided raw material for the development of cellular life. If this model were true for all viruses in the world today, it would mean that they all evolved from a few viral ancestors. In some ways, that would make identifying new viruses and developing vaccines or treatments easier—scientists could take what they know about one virus and use it as a starting point to understand their relatives.
The second so-called “escape” or progressive model of viral origins suggests that viruses arise from genetic elements that escape from the genes of larger organisms. Bacteriophages (viruses that infect bacteria), for example, would come from bits of bacterial genetic material. So according to this model, not all viruses come from deep time—that is, early in the immense arc of the earth’s history, before any multicellular life existed.
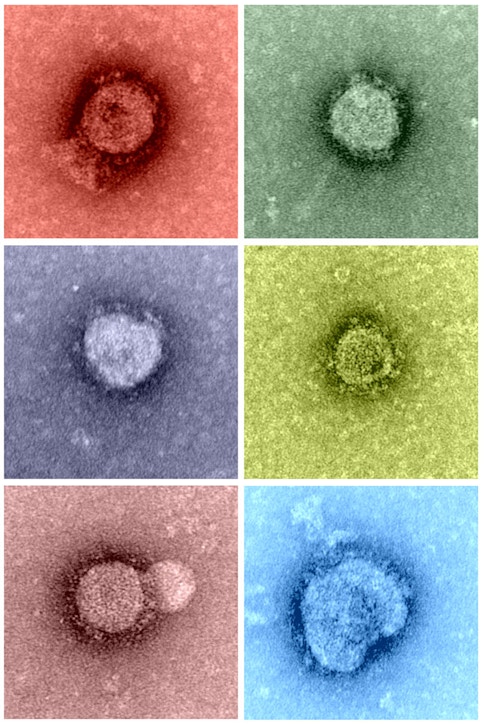
The third model, known as the “reduction” model, is based on a hypothesis that viruses were once larger, free-living organisms. Over time, it is believed that they lost their genetic information and ended up smaller and unable to reproduce alone. But they managed to sustain their existence by replicating in the cells of other organisms. The discovery most commonly interpreted as evidence for this model is that of the mimivirus—the first “giant virus,” which was initially isolated in 2003.
How giant viruses expanded our view of the viral world
Twenty-five years ago, David Wessner, PhD, now a biologist at Davidson College, was a postdoctoral scholar on a research team that was studying coronaviruses. His work involved examining their spike proteins—prongs protruding from the viruses’ exterior—and determining how they interacted with cells. “A lot of that basic research set the stage for the response when COVID-19 emerged,” says Wessner, “and the same could end up being true for basic research on giant viruses. We haven’t yet identified any giant viruses that affect human health dramatically, but that could change.”
What would later be named a mimivirus was first observed during an investigation of a 1992 pneumonia outbreak in England. Bernard La Scola, PhD, a microbiologist at Aix-Marseille Université in France, led a multiyear effort to isolate the unknown entity and examine its structure. He had a friend who at the time was working with electron microscopy. “I gave him a sample,” says La Scola. “I asked him if he could look at this curious matter and told him that I had no idea what it was.’”
La Scola says that when he first saw the resulting image, he wasn’t thinking about a giant virus.
“It was only after I looked at the scale that I realized it was too big to be a virus. And so we began our study with that.” La Scola and his colleagues knew they had isolated something new.
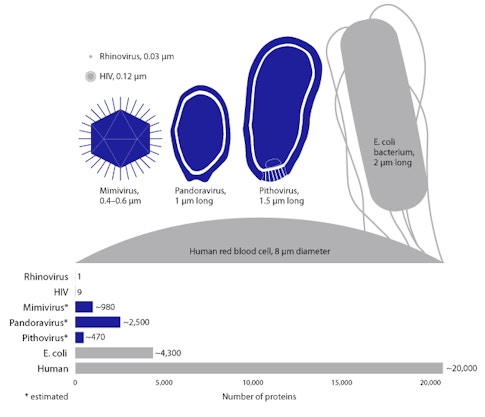
Ultimately, the discovery of the mimivirus (Acanthamoeba polyphaga)—and later the mamavirus and then the even larger pandoravirus—stretched the human perception of what it means to be a virus, given their relative size and genetic complexity. Some scientists think these “giruses” may be the descendants of cells from deep time, while others suggest that they may constitute a fourth dimension of life, distinct from the archaea, bacteria, and eukaryote domains that make up the tree of life model.
La Scola doesn’t engage much in the debates regarding the origins of viruses in deep time; he focuses instead on isolating and identifying new viruses. Over the years, he and his students have received funding from various sources, including his university, the Méditerranée Infection Foundation, and the French Ministry of Health, but La Scola says most of his work on giant viruses has been unfunded. It has benefited from the funding he’s received for other, more applied projects—and from his and others’ keen interest and persistence.
Today, though giant viruses aren’t known to affect human health dramatically, La Scola points warily to Asfivirus, the African swine fever virus. It transmits from ticks to pigs and between pigs. It causes hemorrhagic fever and is closely related to a giant virus that La Scola and his team isolated in Marseille from sewage.
“It’s an example of genome reduction, because it lost about two-thirds of its genome,” he says. “And now this giant virus is probably the agent of hemorrhagic fever in vertebrates—the pigs.” The virus may have come from a larger organism and evolved to cause illness. It happened once, so it could happen again—perhaps in people.
In 2020, La Scola and his colleagues published a paper on the puzzling origins of another novel virus—the Yaravirus. When they sequenced the virus to decipher its genetic information, they realized that some of the genes looked similar to those found in giant viruses, but they couldn’t figure out how or whether the Yaravirus was related to them. None of its genes matched any sequences of known organisms. It was completely new—a virus of dark matter, characterized by unique genetic information. Such dark matter, a vast abyss, is evidence of a viral world we must come to understand better.
The role of human behavior
In intact ecosystems like healthy forests, viruses and their wild hosts have long interacted in a delicate balance with each other. When humans intrude on what scientists call an “existing disease ecology network,” they often expose themselves to unfamiliar viruses. Similarly, taking wild animals out of a forest and transporting them to commercial markets also exposes humans to novel viruses. Looking ahead, humans’ entanglement with the viral world will be shaped in part by “the edge”—that is, the degraded habitats where people and wildlife interact and exchange pathogens.
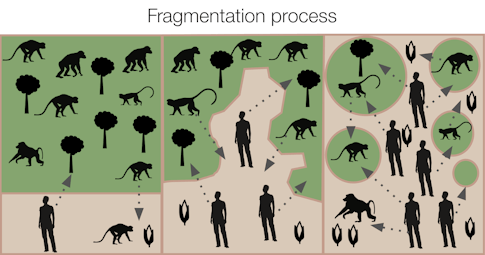
“There’s a related metric: edge density,” says Bloomfield. Edge density refers to the shape and extent of fragmentation in a landscape where humans carry out their daily activities. Cutting trees, building roads, and expanding urban settlements are all activities that can cause such fragmentation.
Environmental degradation creates more “edges” between humans and animal habitats and thus more possibilities that people may come into contact with viruses to which they previously haven’t been exposed. Bloomfield studies how modifying landscapes encourages interactions between people and wildlife and affects infectious disease emergence. Infectious disease doctors often focus on specific diseases, while public health experts commonly pursue the details of spillover events—when a virus moves from one species to another, such as from a bat or monkey to a human. But Bloomfield is interested in much larger patterns: how human behavior drives both connectivity and environmental degradation, increasing edge density and therefore the risk of disease transmission.
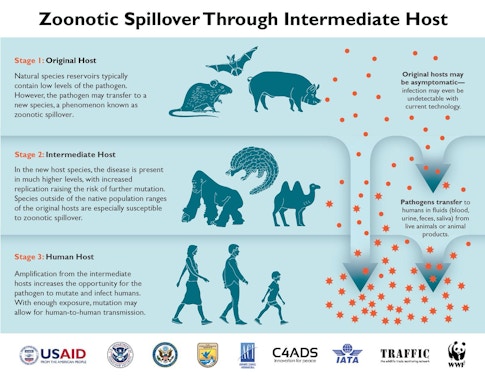
The fishbone pattern of roads cut into the Amazon forest is a common example. In 2000 and 2001, researchers in Peru documented a 278-fold higher biting rate by mosquitos in deforested areas, compared with relatively pristine sites. Cases of malaria rose as mosquitos reproduced quickly in newly sunlit pools and people encroached into the forest. In Uganda, Bloomfield’s research revealed greater edge density around households and activities such as collecting materials for house construction. Foraging in forested habitats also increases the likelihood of contact between people and other primates. Researchers have found that forest degradation has been associated with outbreaks of many viruses that originated in wildlife, such as Nipah, Hendra, and Ebola.
“Areas with higher biodiversity and human encroachment on that biodiversity are definitely areas of concern,” says Bloomfield. “The more interactions between species you have, the higher likelihood you have for rare transmission events,” she notes. Science journalist David Quammen, who wrote a prescient book about the emergence of novel diseases, explains that the phenomenon is similar to the chance of rolling a pair of dice and getting snake eyes. The odds on one roll are low, but roll the dice often enough and eventually you’ll end up with a pair of ones. Spillovers and outbreaks can occur when the right virus transmits to people under the right circumstances. Modern travel and our current level of human connectivity around the globe means that an outbreak can quickly become a pandemic when an infected person travels from one location to another.
Perhaps we may find the right balance only by developing a better understanding of the viral world and the many ways that viruses continue to shape our lives—by attacking us, by becoming part of us, and by helping us, too. Halting degradation and wildlife trade will also help us keep people apart from novel—and potentially harmful—viruses.
Prior to 2015, data on emerging infectious diseases was not even tracked by G-FINDER, a project that monitors and reports on global funding for research and development on neglected diseases—under the auspices of the global think tank Policy Cures Research and with support from the Bill and Melinda Gates Foundation. A combination of the Ebola epidemic; pandemics like SARS, MERS, and H1N1; and mounting concern regarding global health security raised the issue in the collective consciousness even prior to the emergence of SARS-CoV-2. Yet funding for basic science research on viruses and emerging infectious diseases still lags where many experts feel it should be.
Increasing recognition of the interconnections among people, animals, plants, and their shared environment—and the effects of these interconnections on human health—will also require a paradigm shift to more interdisciplinary research and collaboration. Because the health of people, animals, and the environment are deeply intertwined, research efforts and problem-solving require bridges between the efforts of medical and environmental scientists. So far, there aren’t a lot of mechanisms fostering such interdisciplinary research. “I think foundations are probably where we’ll find a lot of this support,” says Bloomfield.
Scientists around the world made history this year by developing an effective vaccine against COVID-19 less than a year after those first images of the novel coronavirus were available. It was basic science research that laid the foundation for such an unprecedented response; in the past, vaccines have often taken 10 to 15 years to develop. Shaped in part by our continued interactions with nature and each other, our future demands a continuation of that approach: putting existing knowledge into action and supporting basic research aimed at deciphering a viral world that we still do not wholly know.
About the Author
Lauren E. Oakes is Conservation Scientist at the Wildlife Conservation Society, an Adjunct Professor in the Department of Earth System Science at Stanford University, and a science writer. She is the author of In Search of the Canary Tree (Basic Books, 2018).
This article first appeared as part of the Science Philanthropy Alliance‘s COVID-19 Basic Science Prequels series. Decades before COVID-19 gripped the world, basic science researchers were laying the groundwork for lifesaving medical and technological breakthroughs now central to the world’s pandemic response. With support from the Kavli Foundation and the Simons Foundation, the Science Philanthropy Alliance has enlisted a team of science writers to explore these science origin stories. Each month, the COVID-19 Basic Science Prequels will unpack the people, history, and serendipitous discovery behind topics that now dominate our daily lives.