Putting the Code in Scientific Computation

During his childhood, Robert Blackwell taught himself computational basics by tinkering with the family computer. While installing new operating systems to test the limits of what the computer could do, Blackwell would often break the computer — sometimes losing digital family photos and other documents in the process — and would then have to figure out how to get it running properly again.
Blackwell’s skills have come a long way since then. He is now a software engineer at the Flatiron Institute’s Scientific Computing Core, or SCC, working with scientists to write and improve their scientific computing programs. He also runs educational seminars and workshops to help researchers at the institute improve their skills and become ambassadors in their fields for better scientific computing practices.
Prior to joining the Flatiron Institute, Blackwell worked as a postdoctoral fellow in biophysics at the Friedrich Alexander University of Erlangen-Nuremberg in Germany. He has a bachelor’s degree in physics from the University of Georgia and a doctorate in physics, with a focus on computational biophysics, from the University of Colorado.
Blackwell recently spoke to the Simons Foundation about his work. The conversation has been edited for clarity.
What kind of work do you do at the SCC?
A good chunk of my work is helping scientists build and write their software to run for supercomputing environments. For example, if a scientist’s code isn’t running well or it’s taking too long to run, I’ll help optimize it. My background in biophysics, computational programming and computer systems administration enables me to fill this niche; I can help scientists both on the computer support side and on the scientific side.
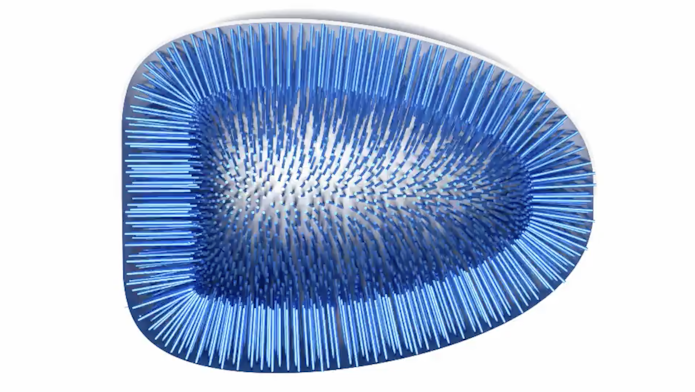
I’m also the core developer on SkellySim, a new biophysics package that simulates motility in the components of biological cells. Cells across pretty much all forms of life have common components, but little is known about how those components physically interact. That’s because their interactions involve a lot of fluid dynamics — the physics that describes how liquids and gases flow. Since fluid dynamics is generally very hard to calculate, it’s often ignored.

SkellySim, however, calculates the effects of fluid dynamics while also simulating some of the more common components of the cell in a very rigorous way. There are not many approximations, which is typically how fluid dynamics are treated, if at all. The math behind SkellySim is very complex, so I work closely with scientists. They provide the mathematics which I code into the program.We think SkellySim will advance what we know about how cellular components interact and move. In some initial tests, we’ve shown that even the simplest system exhibits very different behavior when you add in fluid dynamics. In a system without it, the particles only interact with themselves and anything they touch. But with fluid dynamics, everything moves around and there’s different collective behaviors that affect the interactions of objects that never physically touch.
After two years working on this project, the simulation is just now starting to be used by other scientists at the Flatiron Institute, especially scientists in the biophysical modeling group at the Center for Computational Biology, and we’re publishing some papers about it soon. It still has some work before it’s ready for public use, but once that’s done, we want to advertise it to the larger scientific community. A lot of scientists have already shown interest, and I think once they see what it can do, it’ll catch on quickly.
What will scientists be able to study with SkellySim?
One application is studying microtubules, which are protein complexes inside of the cell that are important for transporting materials from one part of the cell to another. Microtubules are also involved in mitosis, which is the main way cells divide and replicate. In mitosis, microtubules help align and split the DNA. Understanding how microtubules work can help us understand the underlying mechanisms that drive mitosis. SkellySim is also useful for understanding other cell components, like actins and other filaments, and cellular motility, including how bacteria swim.
Are there practical applications of understanding cell motility?
On a fundamental level, we don’t really understand how cells work at small scales. Microtubules are many times smaller than the resolution of most microscopes, so modeling is really our only way of understanding how they work individually.On a more practical level, if you understand how things move and how they interact, you can start thinking of ways that you can manipulate them. There’s a somewhat inside joke in biology that any basic study can be applied to cancer. For us, understanding motility could help scientists engineer medicines that ‘swim,’ allowing targeted drug delivery. Understanding mitosis can help scientists study genetic diseases that are caused by issues in cell division or any kind of transport in the cell. Applications down the road could really be anything.
By clicking to watch this video, you agree to our privacy policy.
How can programs like SkellySim help drive scientific progress?
There are so many ways we can use tools like SkellySim to advance scientific understanding. A lot of cutting-edge problems in science these days are just too hard to solve ‘analytically’ — meaning essentially by hand. Software like SkellySim allows us to solve problems by harnessing the power of computers. From this we can make predictions to drive new experiments and discoveries. Even though designing such software can be a lot of work, this method is usually much faster than trying to tackle problems from a theoretical angle, which involves a lot of trial and error.
Here at the SCC, we’re lucky to have dedicated computer engineering staff who know the science, but whose focus is on software. This allows us to create better, more specialized software that can be supported and shared properly. By taking on some of the computer programming burden ourselves, the scientists can focus on publishing papers and analyzing new results. We of course try to ‘teach a person to fish’ as much as possible, but we can also implement more advanced techniques that are well beyond what scientists can be reasonably expected to master.